15
Sequencing PCR Products
The products of Polymerase Chain Reaction (PCR) can have structures which
make them difficult to sequence. One of the most common problems associated
with sequencing of PCR products is the presence of stops or BAFLs, where the
sequence pauses or stops at artifactual ends in the template (actually the ends
of truncated PCR product). This kit incorporates label by way of a radiolabeled
dideoxy terminator so that only the fragments which were properly terminated
are visible in the sequence. No labeled bands are formed at ‘ends’ in the
template, eliminating many of these artifacts and enabling sequences to extend
to essentially the last base of a PCR product. Artifacts caused by appearance of
double-stranded PCR product on denaturing gels are similarly eliminated since
they are not labeled. Following is information which should assist in producing
high quality, reliable sequence information even with PCR product templates
which have been very difficult to sequence with standard methods.
It is essential that PCR products are of high quality and quantity in order to
obtain high quality sequence information. Problems with high background, low
signal intensity and ambiguities can often be traced to the PCR step. Not every
PCR will yield a product which can be sequenced. Analysis of the PCR product
on agarose gels and optimization of the PCR may be necessary to obtain
quality sequences.
Enzymatic pre-treatment of PCR products
The key step in this method for sequencing PCR products consists of treating
the PCR product with a combination of Exonuclease I and Shrimp Alkaline
Phosphatase
∞
to eliminate any primer or dNTPs which were not incorporated
into the PCR product. These enzymes are available from USB in a reagent pack
(70995) or pre-mixed (ExoSAP-IT™, 78200) with detailed protocols for their
usage. It is recommended that this enzymatic clean-up of the PCR product be
used with this sequencing method.
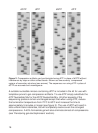
Elimination of compressions
Some DNA sequences, especially those with dyad symmetries containing dG
and dC residues, are not fully denatured during electrophoresis. When this
occurs, the regular pattern of migration of DNA fragments is interrupted; bands
are spaced closer than normal (compressed together) or sometimes farther
apart than normal and sequence information is lost. The substitution of a
nucleotide analog (dITP) for dGTP which forms weaker secondary structure has
been successful in eliminating most of these gel artifacts (18, 19). Two
examples are shown in figure 2 in the sequences run with dGTP.