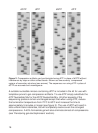
6
Chain termination sequencing
This kit is designed to eliminate sequencing artifacts such as stops (or BAFLs—
bands across four lanes) and background bands. BAFLs can result from the
enzyme pausing at regions of secondary structures in GC-rich templates,
producing prematurely aborted primer extension products of the same length in
all four termination reactions. Background bands can be caused by primer
extensions aborting prematurely at random positions, such as when a template
is rich in a certain base and the complementary nucleotide in the reaction
becomes depleted.
Traditional chain termination sequencing methods (1) involve the synthesis of a
DNA strand by a DNA polymerase
in vitro
using a single-stranded DNA
template. Synthesis is initiated at the site where a primer anneals to the
template. Elongation of the 3' end of the annealed primer is catalyzed by a DNA
polymerase in the presence of 2'-deoxynucleoside-5'-triphosphates (dNTPs),
and is terminated by the incorporation of a 2',3'-dideoxynucleoside-5'-
triphosphate nucleotide analog (ddNTP) that will not support continued DNA
elongation (hence the name ‘chain termination’). Four separate reactions, each
with a different ddNTP, (ddG, ddA, ddT, or ddC), give complete sequence
information. A radiolabeled dNTP (2,3) or primer is normally included in the
synthesis, so the labeled chains of various lengths can be visualized after
separation by high-resolution gel electrophoresis (4,5). In this kit, a radioactive
label is incorporated into the sequencing reaction products at the 3' end by the
use of an [α-
33
P]ddNTP, thus ensuring that only properly terminated DNA
strands are labeled and are visible in the sequence. This results in a cleaner,
more reliable and easier to read sequence with fewer background bands and
virtually no BAFLs.
The accuracy and readability of the sequence obtained depends strongly on the
properties of the polymerase used for chain termination. Some polymerases,
such as Sequenase™ Version 2.0 DNA polymerase, generate much more
uniform, readable bands than others like Klenow and
Taq
DNA polymerase
(6,7,8). Thermostable polymerases, such as
Taq
polymerase, can be used for
multiple rounds (cycles) of DNA synthesis, generating stronger signals. Tabor
and Richardson (9) have discovered that DNA polymerases can be modified to
accept dideoxynucleotides as readily as the normal deoxynucleotide substrates.
Using this technology, a new DNA polymerase for DNA sequencing was
developed. This enzyme, called Thermo Sequenase DNA polymerase, is
thermostable and possesses many of the excellent DNA sequencing qualities of
Sequenase DNA polymerase. The properties of this DNA polymerase include
activity at high temperature and absence of associated exonuclease activity.
Like Sequenase DNA polymerase, derived from T7 bacteriophage, it readily